Trans-PCO: to detect trans-eQTLs of cellular pathways and gene networks
If any figures don’t show, try opening this website in Safari.
Updates
Our paper has been online! See bioarxiv preprint and the site with downloadable results.
Introduction
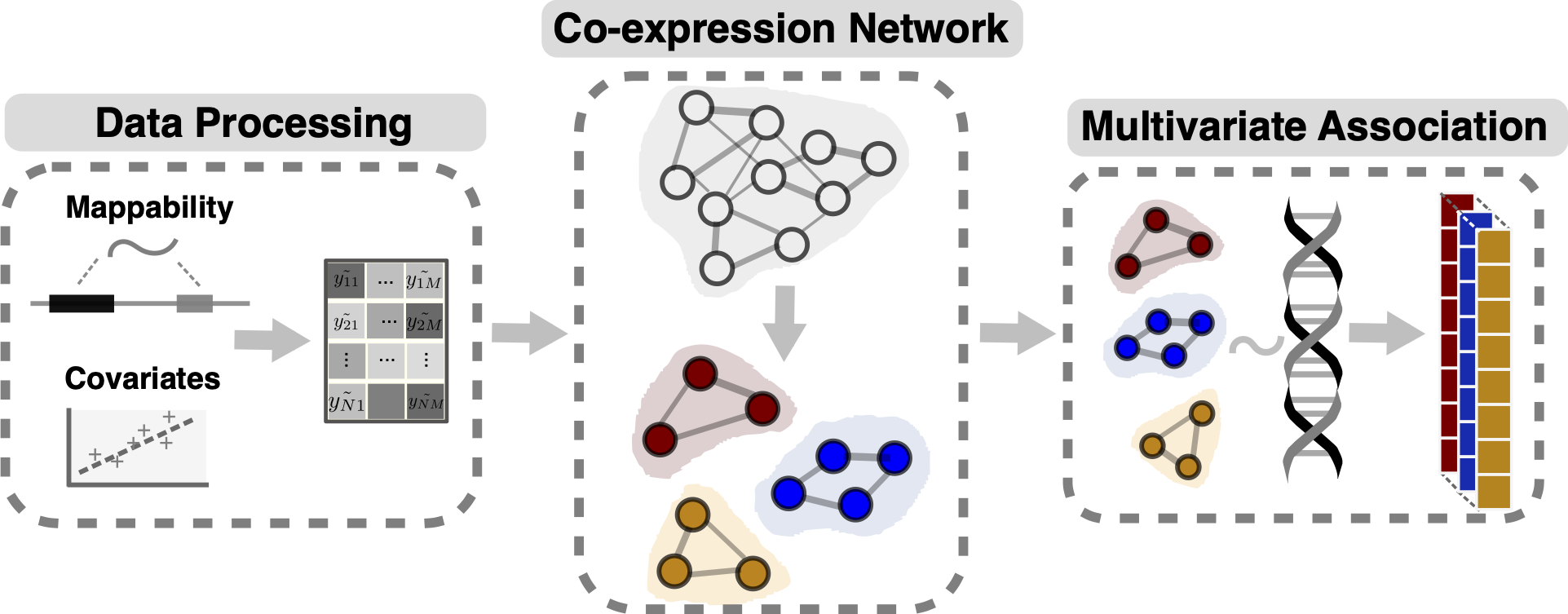
We developed the trans-PCO pipeline to detect trans-eQTLs that are associated with the expression levels of a group of genes (gene module) by using a PC-based multivariate association test [@liu2019geometric] that combines multiple gene expression PCs.
Trans-PCO allows the use of many types of gene groups or sets. For example, genes with correlated expression levels in a co-expression gene network, or genes in the same pathway, or protein-protein interaction network.
Weekly updates
I also host a website to keep track of my weekly work in progress (a wflowr project).